Abstract
Star-convex shapes arise across bio-microscopy and radiology in the form of nuclei, nodules, metastases, and other units. Existing instance segmentation networks for such structures train on densely labeled instances for each dataset, which requires substantial and often impractical manual annotation effort. Further, significant reengineering or finetuning is needed when presented with new datasets and imaging modalities due to changes in contrast, shape, orientation, resolution, and density. We present AnyStar, a domain-randomized generative model that simulates synthetic training data of blob-like objects with randomized appearance, environments, and imaging physics to train general-purpose star-convex instance segmentation networks. Consequently, networks trained using our generative model do not require annotated images from unseen datasets. A single network trained on our synthesized data segments C. elegans and P. dumerilii nuclei in fluorescence microscopy, mouse cortical nuclei in µCT, zebrafish brain nuclei in EM, and placental cotyledons in human fetal MRI, all without any retraining, finetuning, transfer learning, or domain adaptation.
Motivation
Bio-microscopy and radiology studies are often interested in blob-like star-convex shapes like nuclei and nodules. ML experts can then segment these instances with very high performance given annotated training data. Without training data, experts can pull off clever active and interactive learning and domain adaptation methods.
The key assumption there is that ML expertise, retraining, and annotation are feasible for each new segmentation task. We instead segment all star-convex instances in any 3D dataset without needing any new training data or (re)training.
Training data generation
Core idea: We procedurally generate training samples with diverse appearances, shapes, and imaging physics. A network trained to segment these synthetic images should generalize to unseen real datasets.
Our synthesis pipeline simulates randomly distorted foreground shapes and generates intensities for each region using a Gaussian Mixture Model corrupted by multiplicative Perlin noise to simulate texture. It then uses extreme data augmentation to simulate wildly variable imaging physics as below.
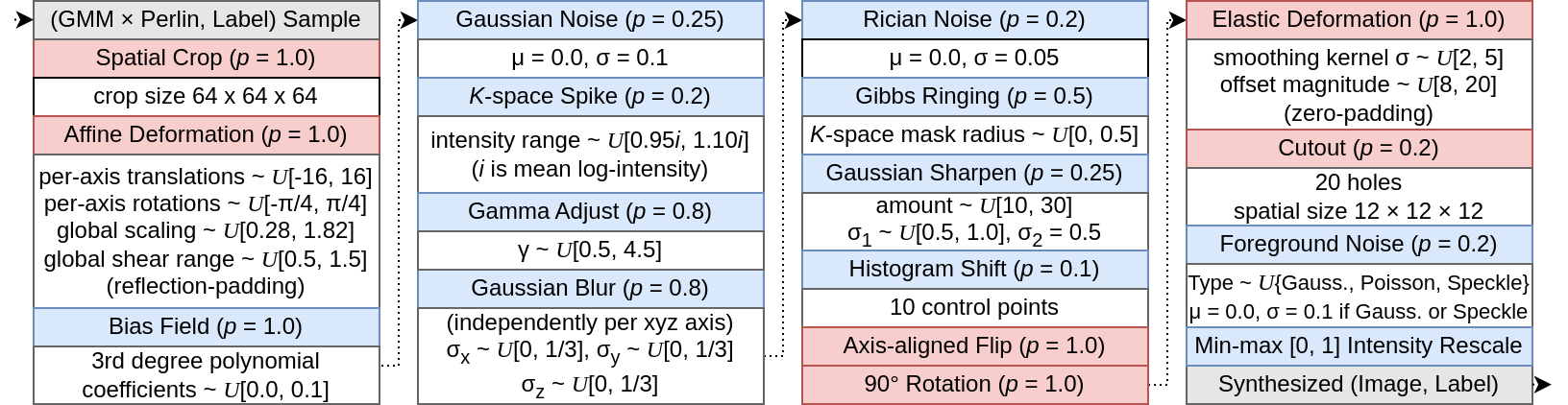
Network training
We train a StarDist network on the synthesized data as it predicts instances that match our shape prior. You could use whichever network you like, though! As below, the trained network learns approximate stability to diverse imaging physics and backgrounds. Try the trained network in Colab.
Caveats
Our (current) method only works for star-convex objects. While that assumption is relevant to several tasks, it won't work on instances that are highly irregular or elongated in shapes. Also, as real data might have wildly variable resolutions and grid sizes, we use sliding window inference that introduces crop size-sensitivity. See best practices here.
Related projects
There's a lot of work that's similar in spirit to ours and that you should also check out.
Domain randomized semantic segmentation. SynthSeg achieves data-agnostic semantic brain segmentation by starting with a training set of brain labels from which to procedurally generate images. As we aim to segment a narrower set of foreground shapes in a broader set of organs and environments, we instead have to synthesize both labels and images.
Large-scale generalist segmentation networks. Other methods using multi-dataset collections of real images have also trained networks that generalize to new settings. For example, Segment Anything (developed concurrently) trained on a billion-scale dataset of real 2D images to achieve impressive interactive 2D segmentation performance on unseen data. We instead focus on the comparatively niche setting of star-convex instances and operate in 3D without requiring any interaction. Similarly, CellPose assembles corpora of real bioimaging datasets to train generalist cell segmentors.
BibTeX
@misc{dey2023anystar,
title={AnyStar: Domain randomized universal star-convex 3D instance segmentation},
author={Neel Dey and S. Mazdak Abulnaga and Benjamin Billot and Esra Abaci Turk and P. Ellen Grant and Adrian V. Dalca and Polina Golland},
year={2023},
eprint={2307.07044},
archivePrefix={arXiv},
primaryClass={cs.CV}
}